| NTL8_UP00577A_1_UniPROBE_shift8 (NTL8:UP00577A_1:UniPROBE) |
 |
0.776 |
0.548 |
6.299 |
0.912 |
0.908 |
10 |
1 |
1 |
11 |
11 |
6.800 |
6 |
; dyads_test_vs_ctrl_m1 versus NTL8_UP00577A_1_UniPROBE (NTL8:UP00577A_1:UniPROBE); m=6/12; ncol2=12; w=12; offset=1; strand=D; shift=8; score= 6.8; --------srTTAAGkAAyc----
; cor=0.776; Ncor=0.548; logoDP=6.299; NsEucl=0.912; NSW=0.908; rcor=10; rNcor=1; rlogoDP=1; rNsEucl=11; rNSW=11; rank_mean=6.800; match_rank=6
a 0 0 0 0 0 0 0 0 17 63 1 0 99 99 1 6 98 97 19 6 0 0 0 0
c 0 0 0 0 0 0 0 0 31 2 17 0 0 1 0 11 0 0 31 62 0 0 0 0
g 0 0 0 0 0 0 0 0 40 35 3 5 1 0 99 39 0 0 13 10 0 0 0 0
t 0 0 0 0 0 0 0 0 12 0 79 95 0 0 0 44 2 3 37 22 0 0 0 0
|
| AT5G60130.ampDAP_M0008_AthalianaCistrome_rc_shift0 (AT5G60130.ampDAP:M0008:AthalianaCistrome_rc) |
 |
0.829 |
0.484 |
0.883 |
0.932 |
0.935 |
7 |
2 |
12 |
7 |
8 |
7.200 |
8 |
; dyads_test_vs_ctrl_m1 versus AT5G60130.ampDAP_M0008_AthalianaCistrome_rc (AT5G60130.ampDAP:M0008:AthalianaCistrome_rc); m=8/12; ncol2=21; w=14; offset=-7; strand=R; shift=0; score= 7.2; tAAGCAAAAhwTAAGcAAAAh---
; cor=0.829; Ncor=0.484; logoDP=0.883; NsEucl=0.932; NSW=0.935; rcor=7; rNcor=2; rlogoDP=12; rNsEucl=7; rNSW=8; rank_mean=7.200; match_rank=8
a 24 124 148 10 8 153 151 135 127 47 87 25 136 143 11 20 155 142 140 128 55 0 0 0
c 12 2 2 0 121 0 1 4 4 41 20 10 3 0 0 87 0 1 1 0 40 0 0 0
g 17 24 3 142 18 0 0 0 0 20 6 12 12 7 142 38 0 3 1 0 12 0 0 0
t 102 5 2 3 8 2 3 16 24 47 42 108 4 5 2 10 0 9 13 27 48 0 0 0
|
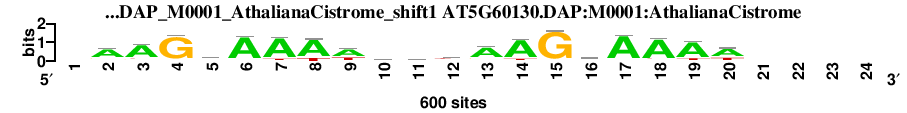
| AT5G60130.DAP_M0001_AthalianaCistrome_shift1 (AT5G60130.DAP:M0001:AthalianaCistrome) |
 |
0.854 |
0.483 |
3.727 |
0.939 |
0.952 |
4 |
3 |
9 |
2 |
3 |
4.200 |
3 |
; dyads_test_vs_ctrl_m1 versus AT5G60130.DAP_M0001_AthalianaCistrome (AT5G60130.DAP:M0001:AthalianaCistrome); m=3/12; ncol2=19; w=13; offset=-6; strand=D; shift=1; score= 4.2; -AAGcAAAwwmtAAGsAAAw----
; cor=0.854; Ncor=0.483; logoDP=3.727; NsEucl=0.939; NSW=0.952; rcor=4; rNcor=3; rlogoDP=9; rNsEucl=2; rNSW=3; rank_mean=4.200; match_rank=3
a 0 421 464 21 121 532 510 473 376 217 221 149 438 501 13 114 540 517 477 386 0 0 0 0
c 0 31 18 12 276 19 1 0 22 117 159 80 22 6 0 264 24 15 18 24 0 0 0 0
g 0 83 57 538 128 33 24 9 26 82 72 87 84 24 563 155 11 25 11 26 0 0 0 0
t 0 65 61 29 75 16 65 118 176 184 148 284 56 69 24 67 25 43 94 164 0 0 0 0
|
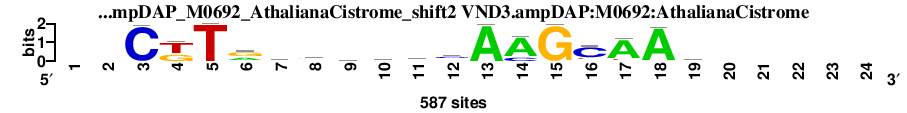
| VND3.ampDAP_M0692_AthalianaCistrome_shift2 (VND3.ampDAP:M0692:AthalianaCistrome) |
 |
0.850 |
0.464 |
5.076 |
0.935 |
0.949 |
5 |
4 |
4 |
4 |
6 |
4.600 |
5 |
; dyads_test_vs_ctrl_m1 versus VND3.ampDAP_M0692_AthalianaCistrome (VND3.ampDAP:M0692:AthalianaCistrome); m=5/12; ncol2=17; w=12; offset=-5; strand=D; shift=2; score= 4.6; --CkTraarwwsAAGCAAy-----
; cor=0.850; Ncor=0.464; logoDP=5.076; NsEucl=0.935; NSW=0.949; rcor=5; rNcor=4; rlogoDP=4; rNsEucl=4; rNSW=6; rank_mean=4.600; match_rank=5
a 0 0 7 0 0 221 236 272 204 197 225 56 584 475 0 35 500 569 97 0 0 0 0 0
c 0 0 580 0 0 42 124 68 109 114 76 295 3 112 0 435 18 0 222 0 0 0 0 0
g 0 0 0 239 0 304 95 146 150 80 104 155 0 0 583 32 14 0 97 0 0 0 0 0
t 0 0 0 348 587 20 132 101 124 196 182 81 0 0 4 85 55 18 171 0 0 0 0 0
|
| dyads_test_vs_ctrl_m1_shift7 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=17; shift=7; ncol=24; -------wwattAAGCAAAtawww
; Alignment reference
a 0 0 0 0 0 0 0 212 196 335 108 88 483 488 20 36 498 478 384 109 254 170 171 191
c 0 0 0 0 0 0 0 69 82 54 40 58 3 15 19 443 12 14 45 37 78 88 100 81
g 0 0 0 0 0 0 0 89 105 50 68 64 11 11 470 15 9 11 38 55 78 69 97 93
t 0 0 0 0 0 0 0 161 148 92 315 321 34 17 22 37 12 28 64 330 121 204 163 166
|




